In situ hybridization (ISH) is a technique used to detect nucleotide sequences in cells, tissue sections, and even whole tissue. This method is based on the complementary binding of nucleotide probes to specific target sequences of DNA or RNA. These probes can be labeled with either radio-, fluorescent-, or antigen-labeled bases. Depending on the probe used, autoradiography, fluorescence microscopy, or immunohistochemistry, respectively, are used for visualization.
Since the first appearance of ISH, many different methods have been developed. Some of the more common methods are discussed below.
Chromogenic In Situ Hybridization (CISH)
CISH uses peroxidase- or alkaline phosphatase-labeled reporter antibodies to hybridize with DNA probes, and a bright-field microscopy is used to detect the enzymatic reaction. With CISH, changes in gene amplification, gene deletion, chromosomal translocations, and chromosomal number can all be viewed at the same time, along with tissue morphology.
Advantages of CISH are the fact that signals do not diminish over time, its low cost, the ability to use a light microscope, and permanent staining. CISH is a suitable alternative to FISH, and is widely used by pathologists because it uses brightfield microscopy.
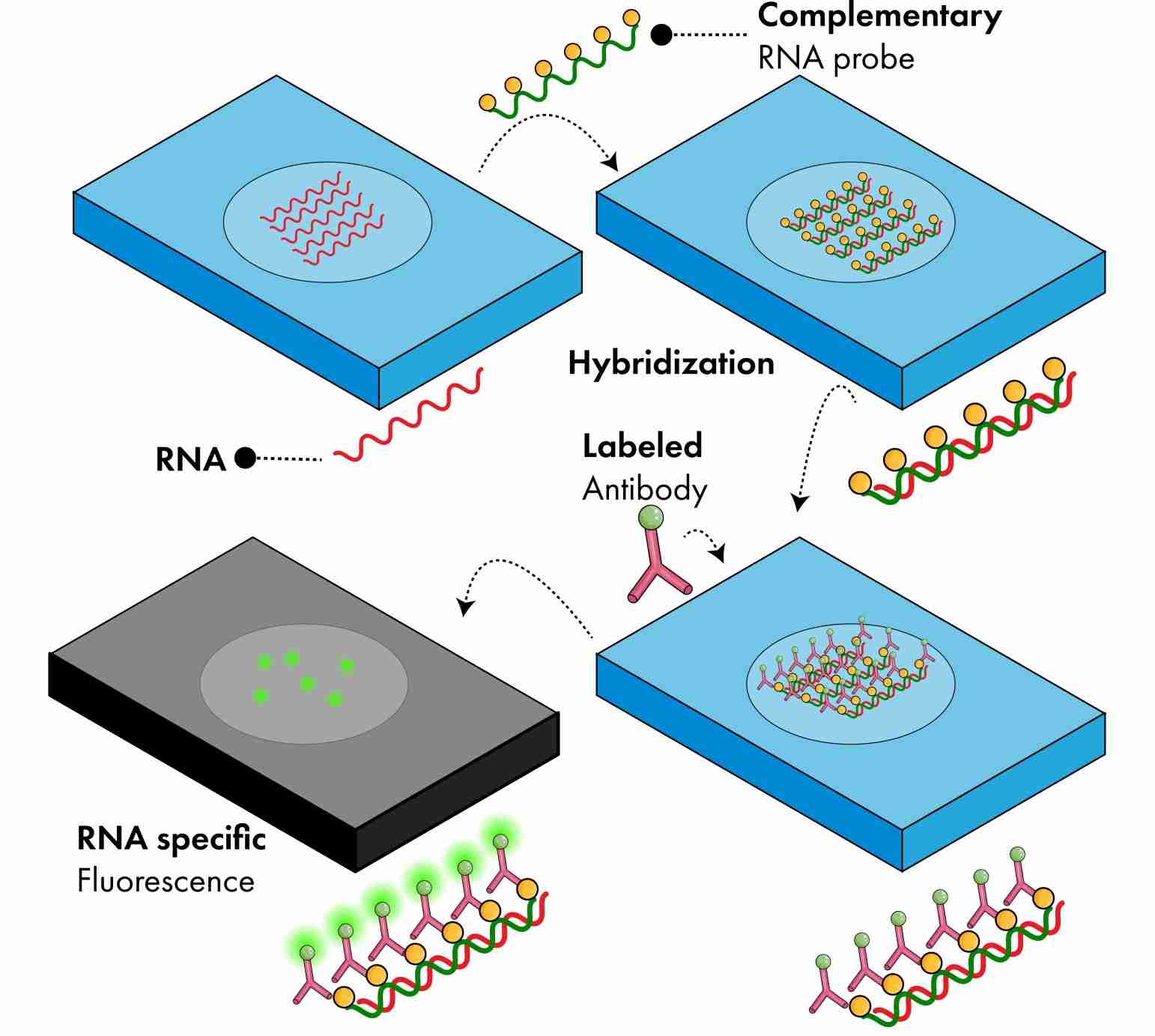
Fluorescence In Situ Hybridization (FISH)
FISH uses directly labeled fluorescent nucleotides or probes with reporter molecules that can be detected indirectly by fluorescent antibodies or affinity molecules. The probes bind to complementary nucleotide sequences to produce fluorescent spots that can be detected in situ using a fluorescent microscope. FISH is commonly used for detection of structural rearrangements such as translocations, inversions, insertions, and microdeletions, as well as for chromosomal gene mapping and for identifying and characterizing chromosome breakpoints.
FISH is highly sensitive and can be used on fresh, frozen, or paraffin-embedded samples, making it a versatile and useful technique for diagnostic applications.
Multicolor FISH (SKY, M-FISH)
In multicolor FISH, two or more probes are each specifically labeled, combined, and then identified with different fluorescent colors. Using this method, scientists can evaluate multiple chromosome sites.
Spectral karyotyping (SKY) and multiplex-FISH (M-FISH) provide the same type of information but the way chromosomal images are acquired and processed is different. SKY identifies the differentiation of the chromosomes based on their spectral properties, whereas M-FISH identifies the differentiation of the chromosomes based on the presence or absence of fluorochrome when visualized with specific filters. The resulting analysis for both methods is the same, revealing hidden translocations and insertions as well as chromosomal components of the marker chromosomes.
The strength of multicolor FISH lies in its ability to define translocations, marker chromosomes and complex rearrangements, and to reveal cryptic changes. However, it cannot detect intrachromosomal rearrangements, such as duplications, very small deletions, or small paracentric inversions.
Comparative Genomic Hybridization (CGH)
CGH is a two-color quantitative FISH method mostly used for investigating copy number variations (CNV) in DNA from two different sources. CGH technique (in comparison to FISH) is less time-consuming and provides an overview of specific chromosomal regions that have undergone deletion or duplication (which causes pathogenesis). It is more efficient in the analysis of oncogenes/cancer.
Array-CGH is the modified CGH technique. It is a chip-based technology that is less than an inch in size. There are thousands of probes in a grid on the chip; differently colored test and control samples are mixed in the grid where they compete to bind with the available probes. The procedure and analysis to be done are the same as CGH. But, this technique (array-CGH) is more efficient than CGH, as it can simultaneously analyze large samples and multiple loci of the chromosomes in very less time.